Segmentation of brain tumor in magnetic resonance images
The dataset used for development was obtained from The Cancer Imaging Archive (TCIA) and involved 110 cases of lower-grade glioma patients. Registers brain MR images with manual FLAIR abnormality segmentation masks are published as a Kaggle Dataset lgg-mri-segmentation.
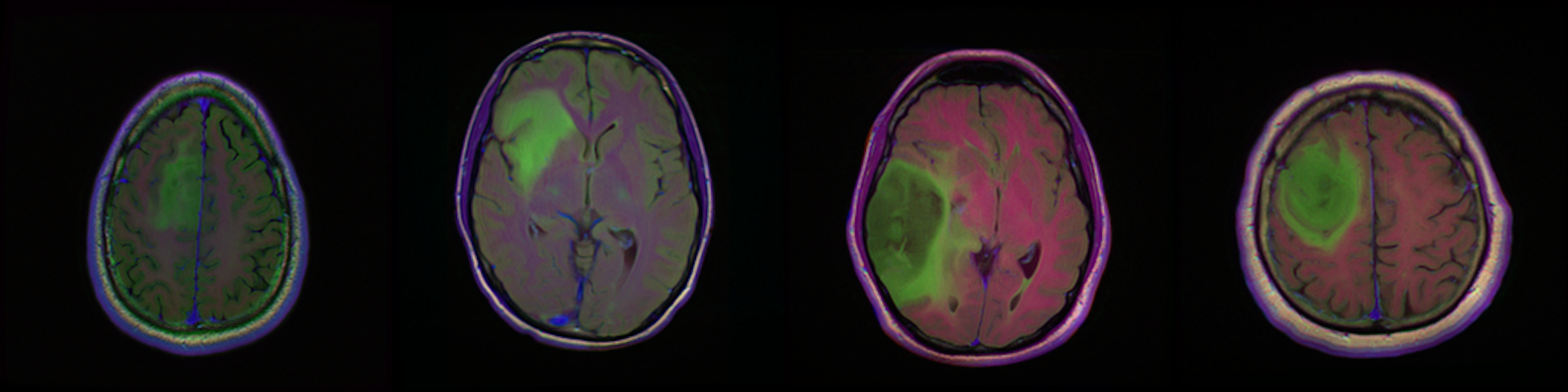
Together with the dataset, we shared a GPU enabled Python code that can be run on Kaggle: kaggle.com/mateuszbuda/brain-segmentation-pytorch. The script implements a data loading, preprocessing, as well as training and inference of a U-Net segmentation model. Trained weights and results can be downloaded form Kaggle.
We applied U-Net architecture for the task of FLAIR abnormality segmentation in brain MRI (Figure 1). Source code for network architecture, training, and inference implemented in PyTorch together with model weights are available on GitHub repository: github.com/mateuszbuda/brain-segmentation-pytorch.
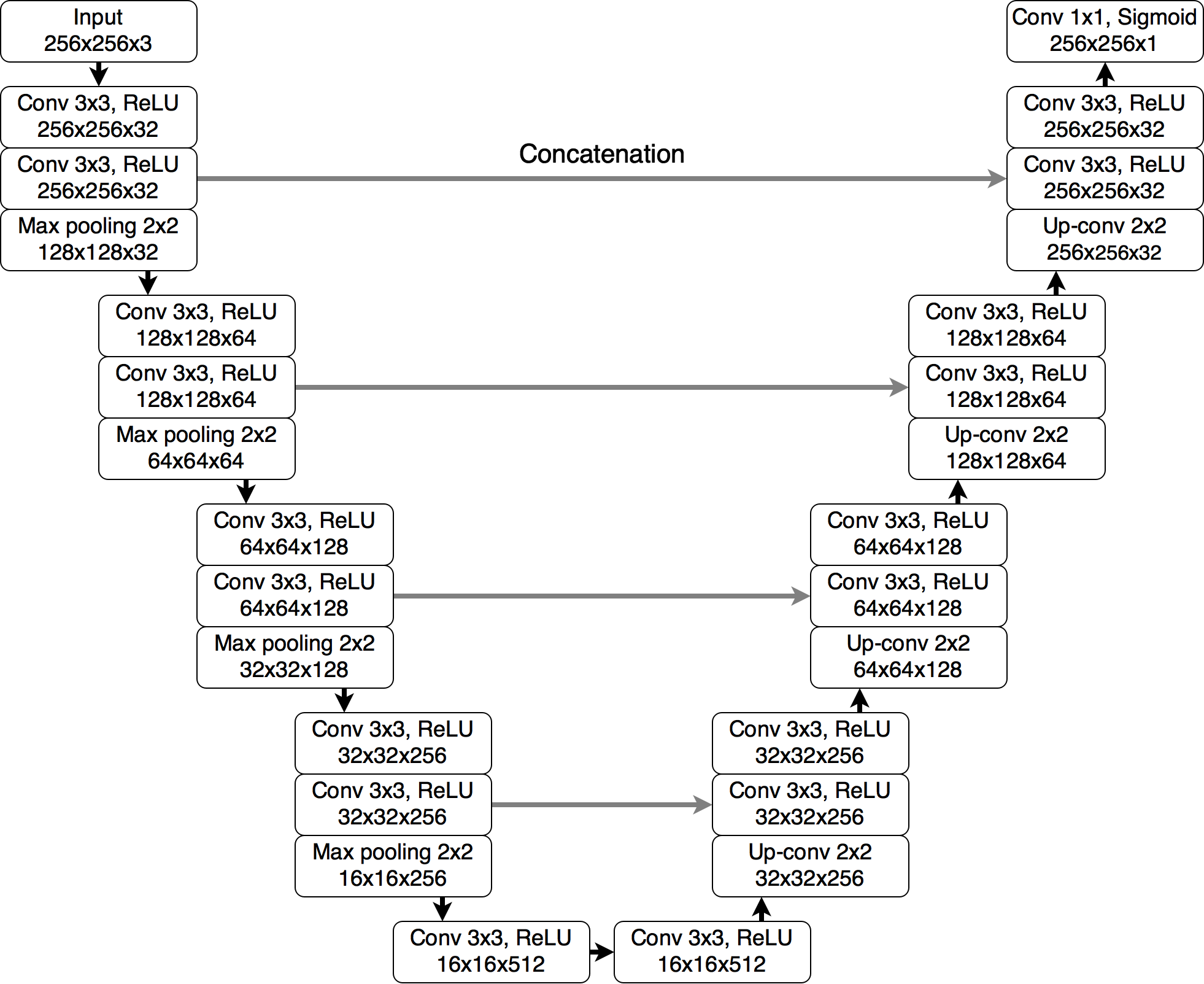
Figure 1. The U-Net architecture used for segmentation.
To evaluate the quality of segmentation, we used Dice similarity coefficient (DSC) with 22-fold cross-validation. The achieved performance was 83.60% mean DSC and 87.33% median DSC. In comparison, DSC of two expert human readers for this kind of tumor is 84% with a standard deviation of 2%. This puts our method on a par with radiologists.
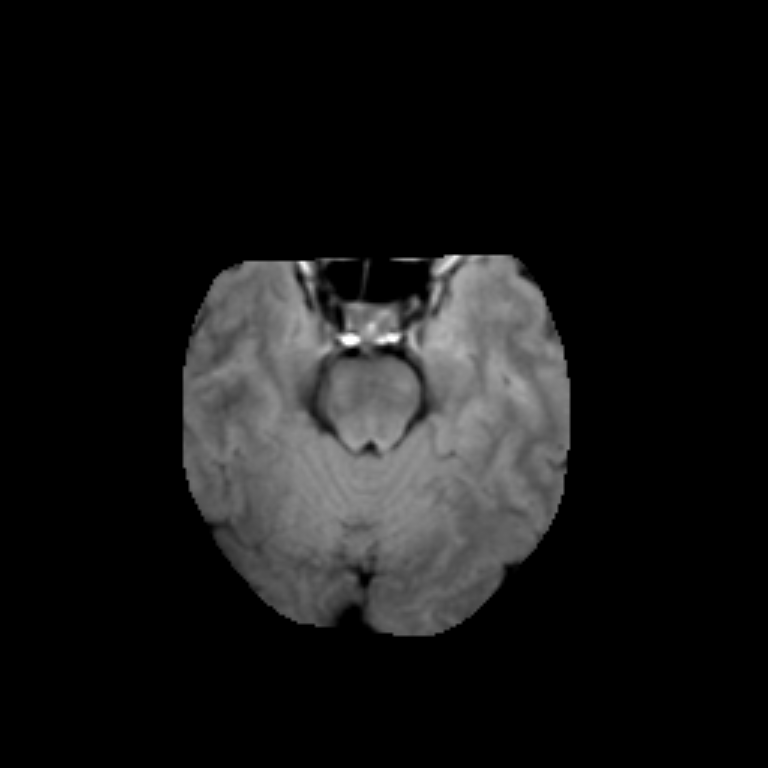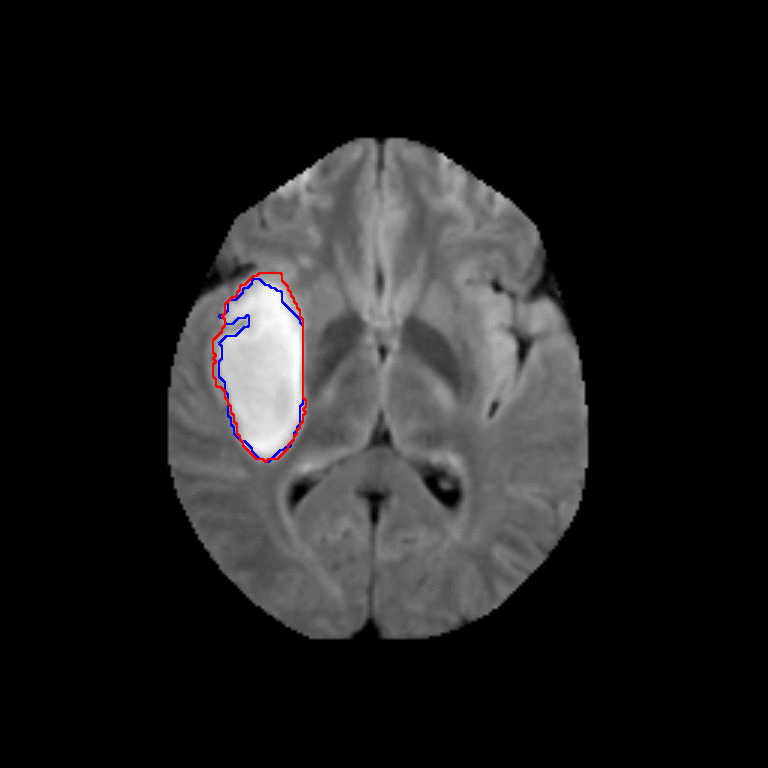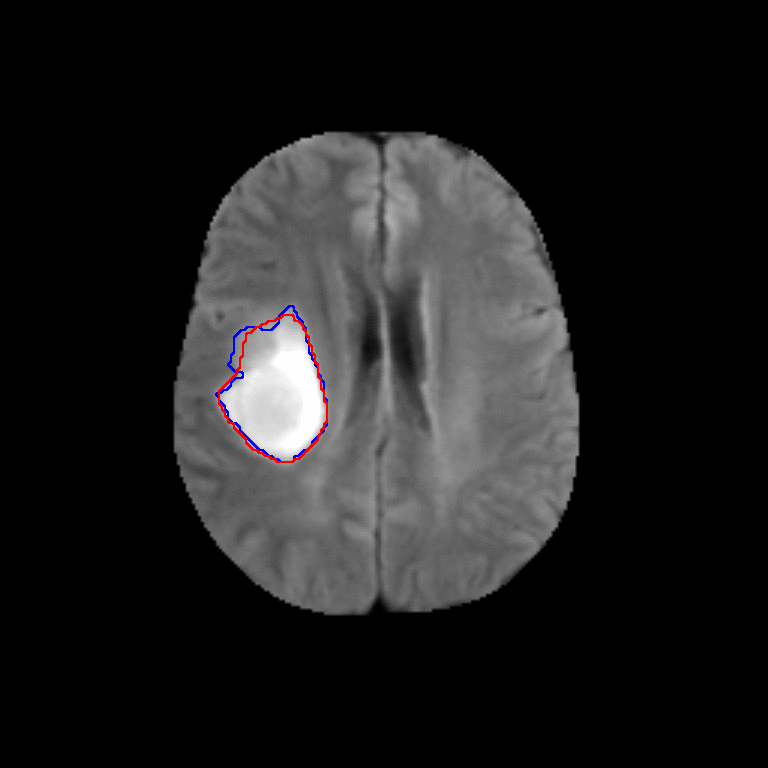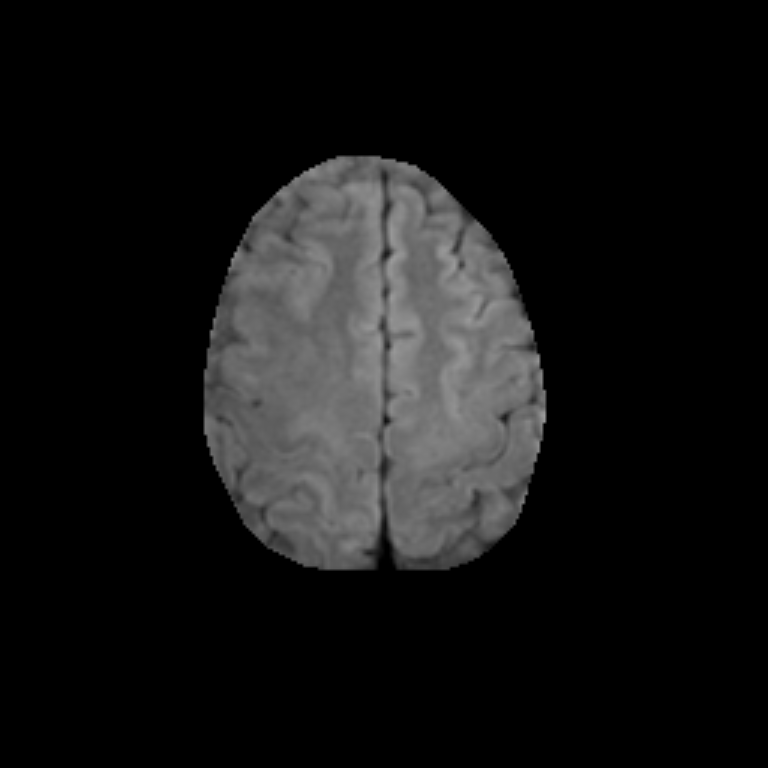
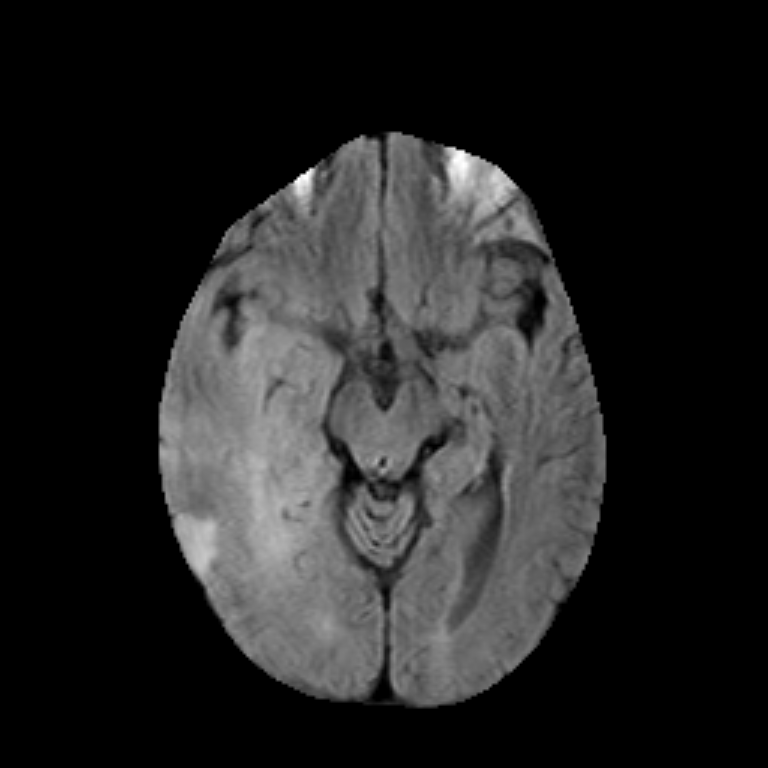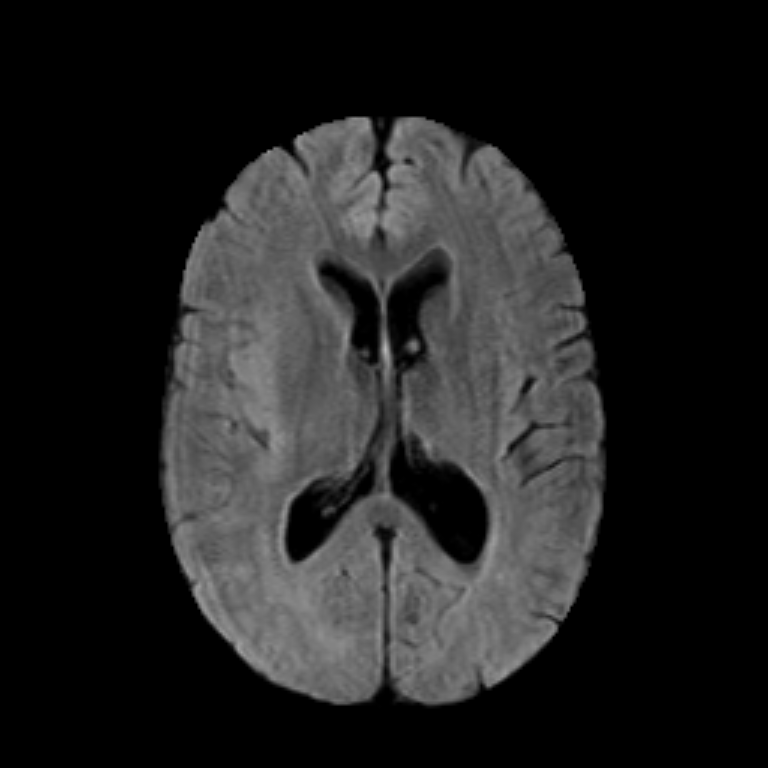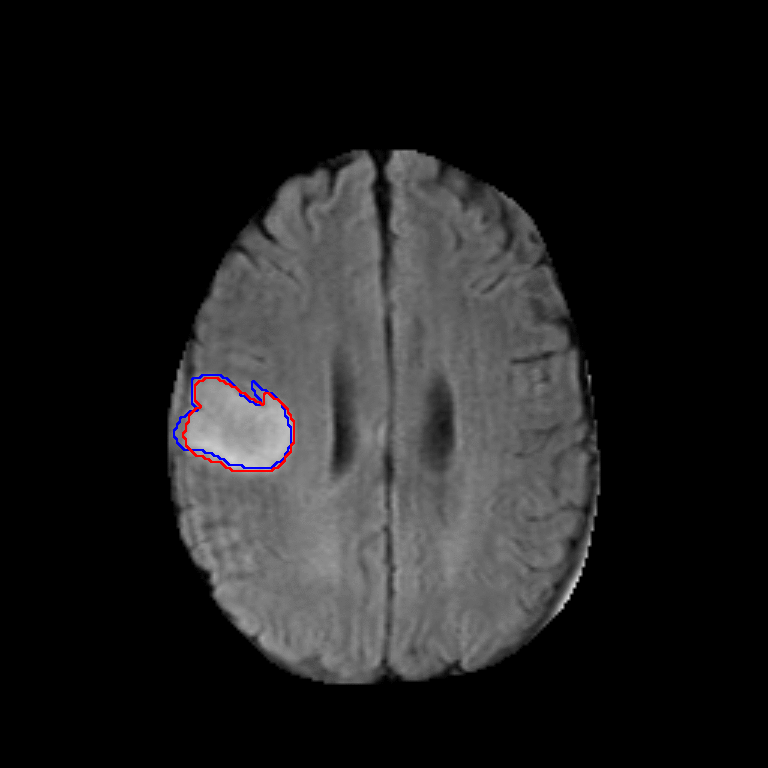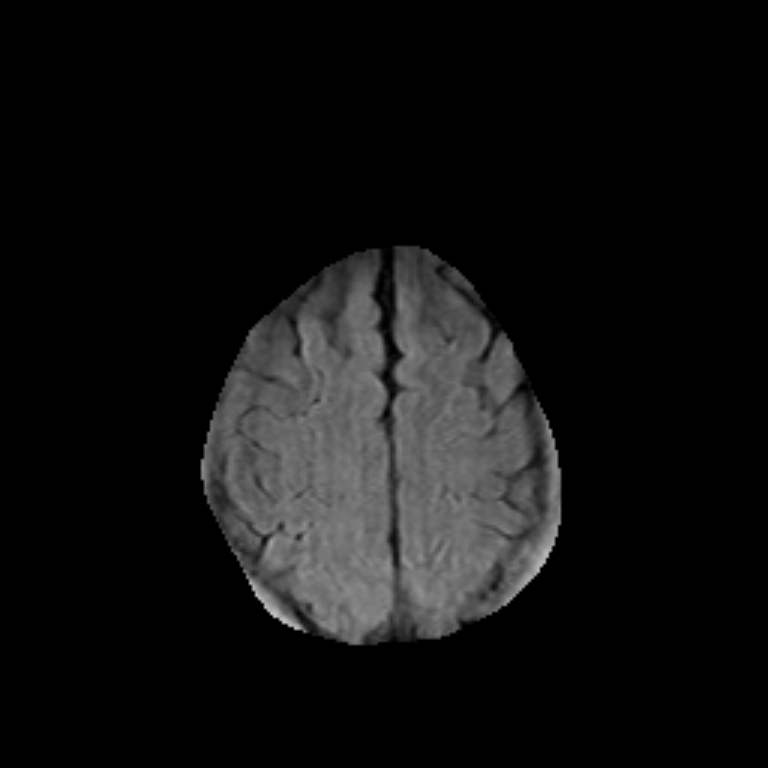
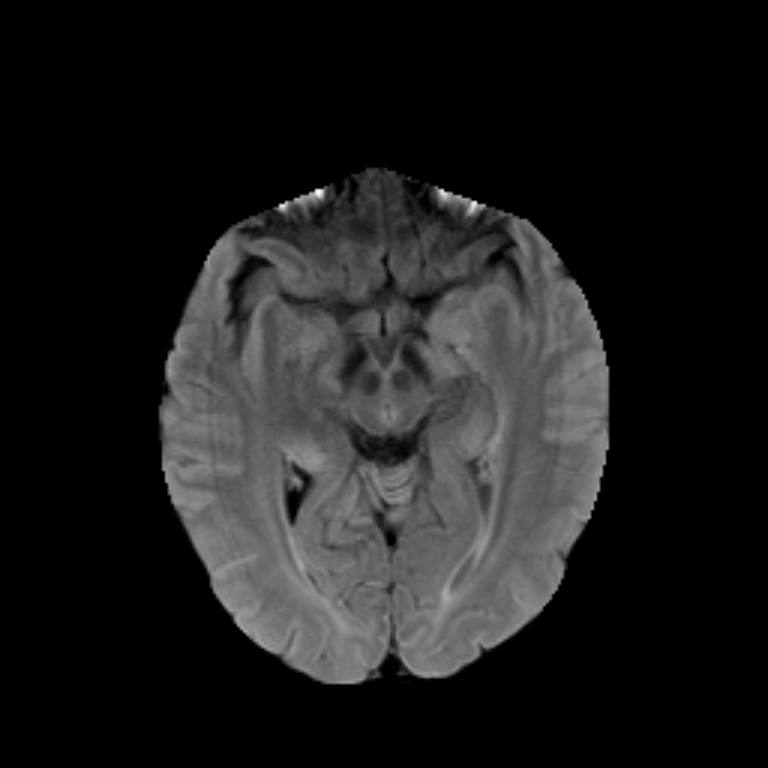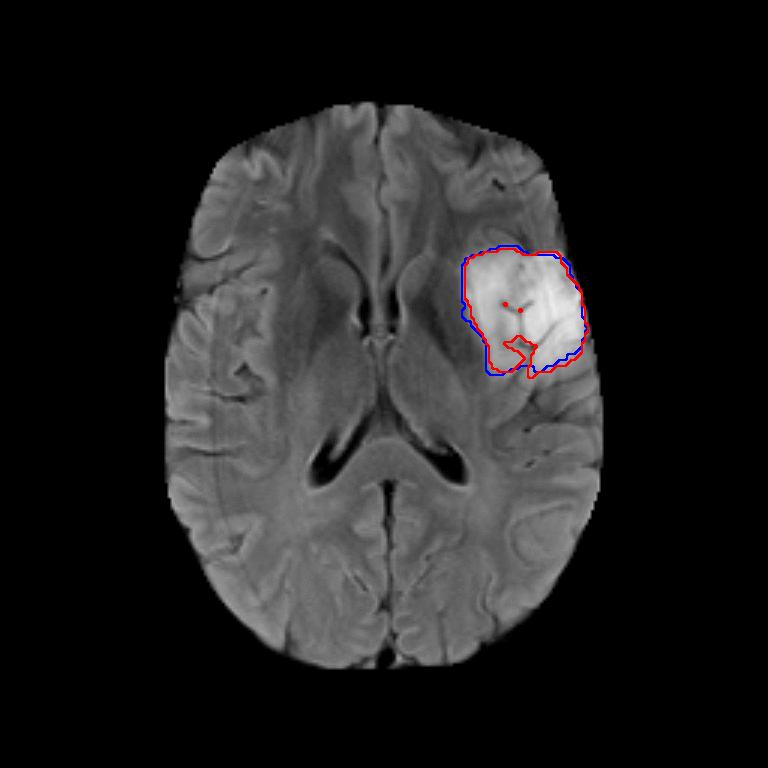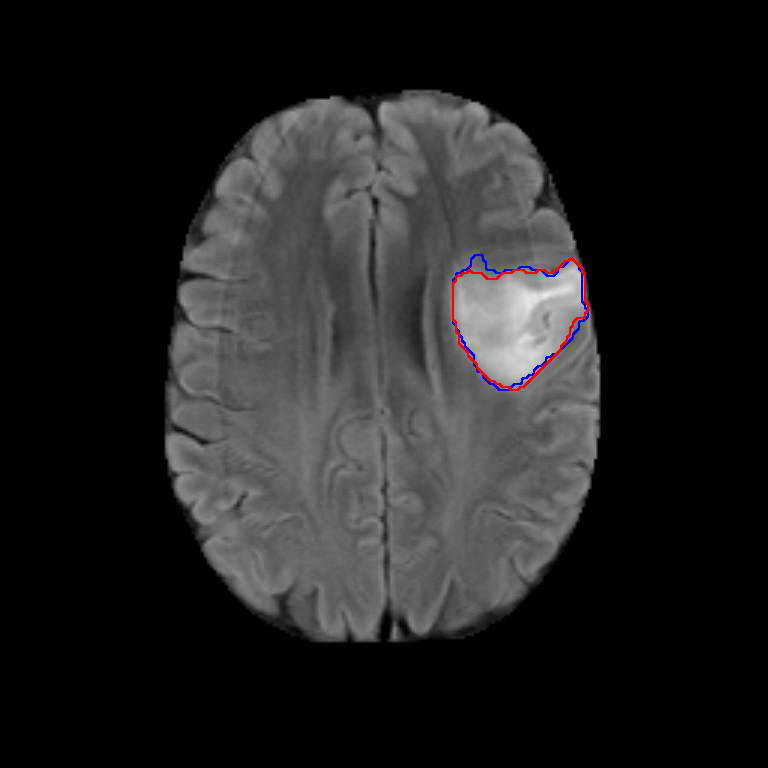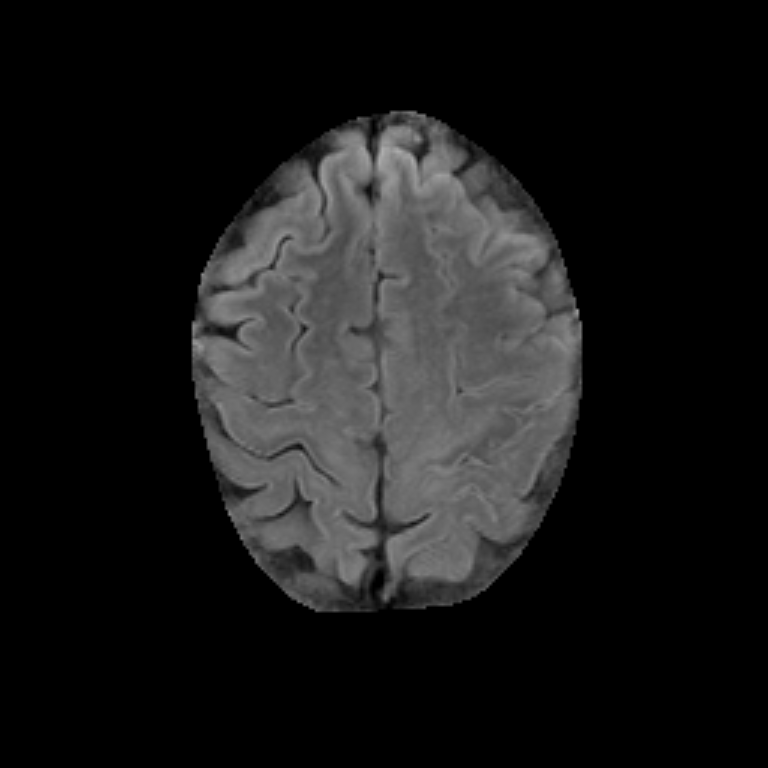
Figure 2. Qualitative results of segmentation. Blue outline corresponds to ground truth and red to automatic segmentation output. Images show FLAIR modality after preprocessing and skull stripping.
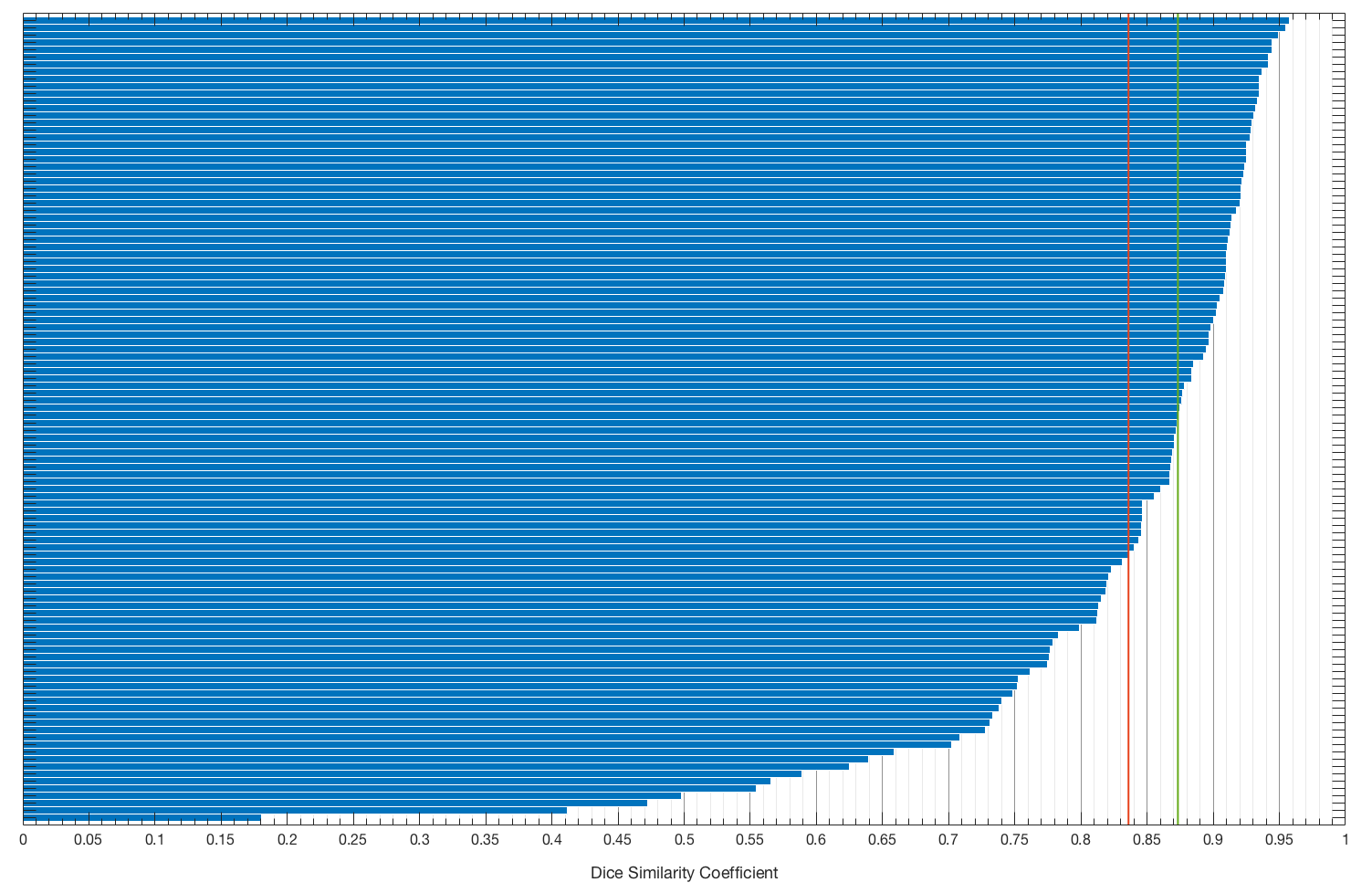
Figure 3. Distribution of Dice similarity coefficient among cases. Red line corresponds to mean DSC and green to median DSC.
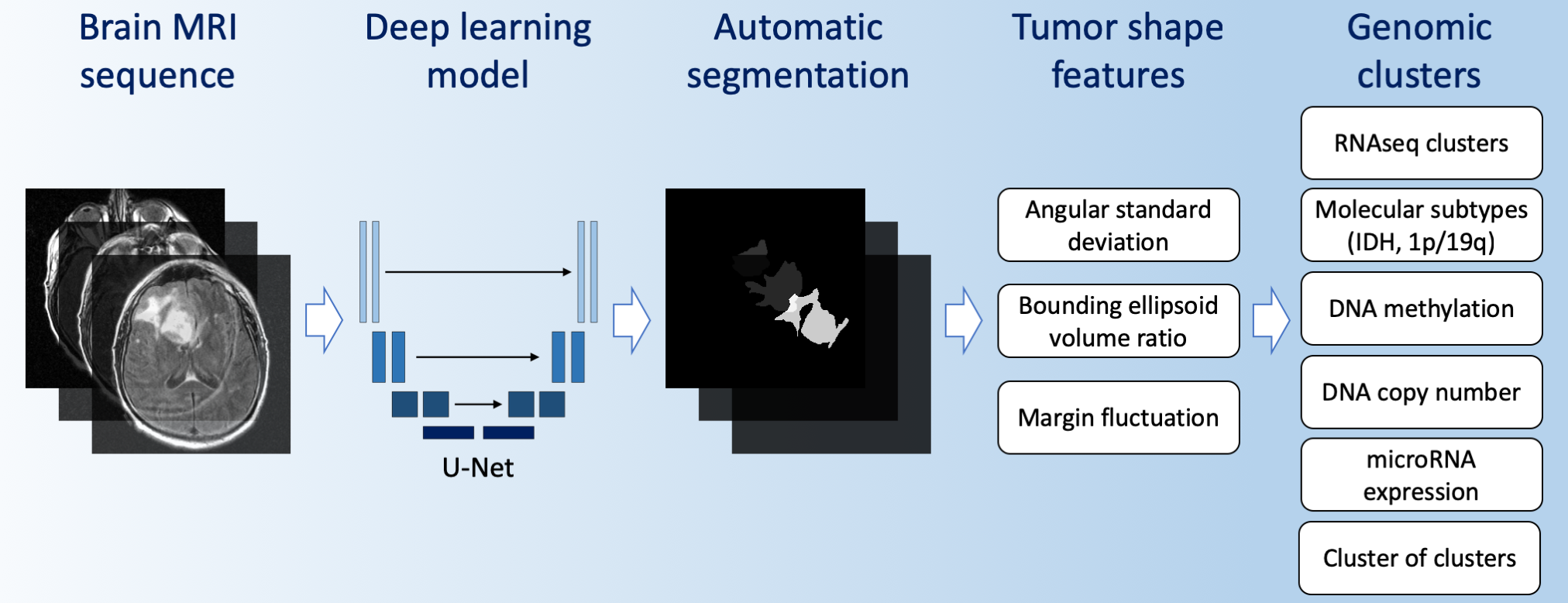
Then, produced segmentations were used to extract 2D and 3D tumor shape features that are predictive of its genomic subtypes. More detailed description of methods and results is available in our paper Association of genomic subtypes of lower-grade gliomas with shape features automatically extracted by a deep learning algorithm published in Computers in Biology and Medicine.
Links
- Full paper: Computers in Biology and Medicine
- Code: github.com/mateuszbuda/brain-segmentation-pytorch
- Data: lgg-mri-segmentation